Use case #4 Predictive (toxicology) modelling workflows
How to integrate data to make predictions.
The context
Toxicological modelling often involves downloading data from multiple sources and subsequent interrogation of their value to existing predictive models or to those being built from scratch. After downloading information from various sources, data to be interrogated (or integrated) is often stored as csv/excel files; unfortunately, such data can be easily corrupted by end-users often without traceability.
The challenge
Integrate and process multiple curated datasets for analyses of DILI (drug induced liver injury) risk.
The solution
EdelweissData™ APIs served the following functions:
- As the source of custom datasets not easily available elsewhere and pre-formatted for predictive modelling;
- To check on data integrity throughout the study by ensuring: use of designated (validated) APIs throughout and traceability of any data changes made through pre-processing steps;
- Provide compliance with FAIR data principles.
We used DILI modelling as an example, but this approach may be replicated for other predictive toxicological/modelling analyses that require interrogation of diverse and heterogeneous datasets in a manner that ensures results integrity/process traceability.
Two datasets available as EdelweissData APIs were compared against each other individually and when combined for their relative performance in Drug Induced Liver Injury/DILI risk prediction for 350 pharmaceutical drugs. Specifically, the ability of these datasets to discriminate “mostDILI” (highest DILI risk) vs “otherDILI” (lower/no DILI risk) drugs was evaluated using logistic regression.
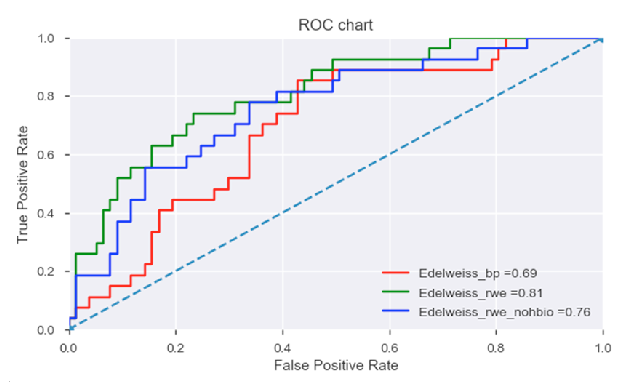
In the above figure, performance of individual datasets is compared (after prior analysis demonstrating superior performance over a combined dataset) using receiver-operating-characteristic (ROC) curve analysis.
As indicated, the Edelweiss_rwe dataset with an AUC value of 0.81 (and a subset of the same, Edelweiss_rwe_nohbi with an AUC value of 0.76) outperformed the benchmark predictors (Edelweiss_bp AUC value of 0.69).