
SciLicium Bioinformatics
Tailor-made bioinformatics analyses for genomics projects
SciLicium is based in Rennes (Brittany, France) and specialized in multi-omics data analysis and the development of workflows and web app solutions in bioinformatics. Over the last few years, SciLicium has also developed strong expertise in toxicogenomics (read-across, adverse outcome pathways, mechanisms of action) including predictive toxicogenomics based on statistical modeling (Machine and Deep Learning).
Determine the toxic potential of compounds is an important challenge faced by scientists and regulatory authorities alike (Grimm, 2011). The genomic-scale data rapid generation has led to the development of toxicogenomics. They combine classical toxicology approaches with (metabol-, prote-, transcript-, epigen-) omics technologies to identify not only deregulated molecular mechanisms upon exposures but also candidate biomarkers for toxicity prediction. In the risk assessment, process transcriptomics datasets (and their resulting gene expression signatures) are now used more and more to:
- (i) characterize the molecular mode of action of chemicals, small molecules, and nanomaterials exposure determined by their toxicogenomics signature (e.g. over/under-expressed genes after exposure to the compound) (Darde et al., 2018)
- (ii) predict the genomic behavior underlying the exposure and its toxicity (Liu et al., 2019)
- (iii) define the adverse outcome pathways (AOP) (Brockmeier et al., 2017)
- (iv) determine the benchmark dose and estimate the critical point of departure for human health risk assessment (Farmahin et al., 2017)
- (v) predict the behavior of uncharacterized compounds by comparing them to other substances whose molecular effects are known (read across) (Pawar et al., 2019)
In this context, SciLicium Toxicogenomics services aim at using transcriptomics data to address several challenges in toxicogenomics. After careful planning of exposure conditions, RNA extraction, and library preparation, the toxicogenomics data are pre-processed and used to identify the toxicogenomics signature, to perform functional analysis, and predict and prioritize potential adverse effects.
Services Overview
SciLicium Toxicogenomics services include:
- A strong expertise in toxicogenomics
- A know-how in toxicogenomics data analysis
- The development of data analysis workflows for standardizing procedures
- The deployment of a dedicated web application for sharing data and communication purposes.
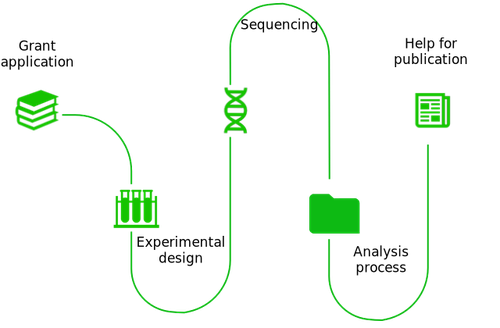
Toxicogenomics expertise
Our experience in a wide range of sequencing technologies (DNA chips, RNA-seq, ChIP-seq, single-cell RNA-seq, 5 'or 3' RNA-seq) makes SciLicium the perfect partners for your projects.
- Experimental design
With its experience in a wide range of technologies, SciLicium can advise you on the optimal experimental and sequencing strategy to choose to acquire the toxicogenomics data set necessary for your project.
- Sequencing
Over the years, SciLicium has created an international collaborators network and makes it available to you for your sequencing project.
- Data management and analysis
SciLicium can get involved at different levels of your analysis: from guidance to help you define your data analysis and data management strategy to a more active role in analyzing your data and/or setting up pipelines and web tools.
Toxicogenomics data analysis
Basic, Standard, or Advanced, it is up to you to choose and customize the analysis that will suit your project. SciLicium’s experience in toxicogenomics data processing and analysis allows us to offer you various benefits on a wide range of technologies (DNA chips, RNA-seq, single-cell RNA-seq, 5 'or 3' RNA-seq) to screen your compounds.
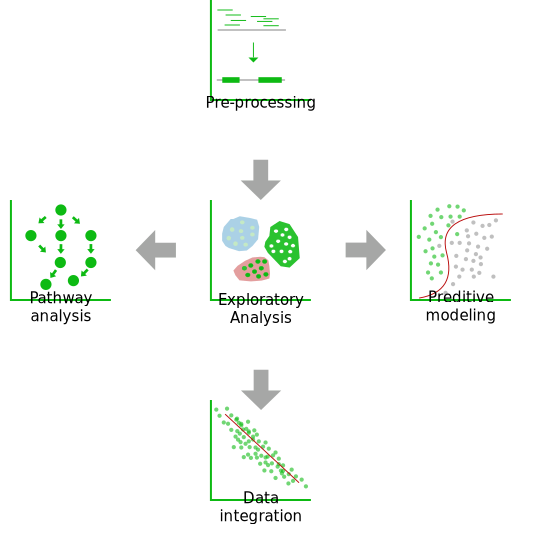
Our Analyses
Basic
You have massive toxicogenomics data and you want to make the best of it without having to worry about the technical aspects or without wasting time on stages such as quality control, data normalization, and basic sample comparisons. SciLicium can take over data preprocessing with standardized and adapted procedures. This critical step will enable you to ensure your samples and sequencing data quality and reliability before starting long and tedious analysis steps.
Standard
After having ensured the high quality of your data, you will want to get to the heart of the matter and answer some of your questions/start your analysis. Unfortunately, you do not have the bioinformatics and biostatistics skills to answer your questions.
Advanced
The “Advanced analysis” service corresponds to a deeper involvement of SciLicium into the exploration and interpretation of your results.
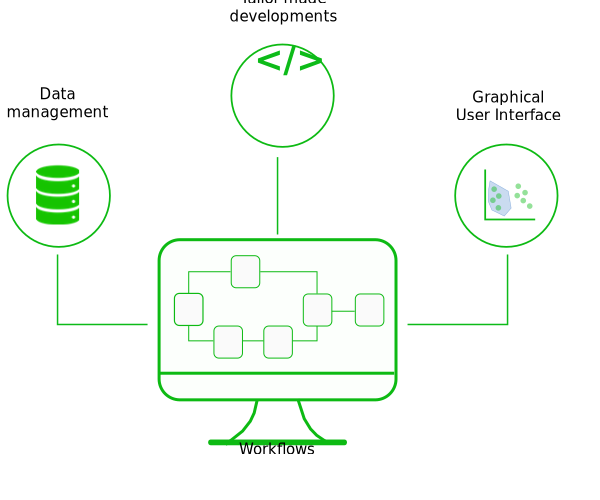
Workflows & web application development
Tailor-made workflows: standardization of your procedures
Automate your data analysis, save time, and increase your study reproducibility while being flexible enough to adapt to your project needs.
Through the various projects carried out by SciLicium, several analysis workflows (DGE-seq, BRB-seq, RNA-seq, single-cell RNA-seq, metagenomics) have been set up and adapted to answer your scientific questions. SciLicium uses the workflow editing standards (Snakemake, Nextflow, CWL) to offer analysis stability over time.
Deploy your bioinformatics tools online
Web application creation is now a major activity in the bioinformatics field in order to share research.
At SciLicium, we have acquired web development skills and expertise to deploy solutions. By working closely with you, we can help you develop innovative bioinformatics web applications with advanced and reliable technologies to meet your project-specific requirements.
SHOP
SciLicium offers a basic, standard, or advanced service for analyzing "-omics" data from different technologies (DNA chip, Single-cell RNA-seq, 3 'or 5' sequencing, metagenomics,. ..) fitting to the needs and scientific question.
- Brockmeier, E. K., Hodges, G., Hutchinson, T. H., Butler, E., Hecker, M., Tollefsen, K. E., Garcia-Reyero, N., Kille, P., Becker, D., Chipman, K., Colbourne, J., Collette, T. W., Cossins, A., Cronin, M., Graystock, P., Gutsell, S., Knapen, D., Katsiadaki, I., Lange, A., … Falciani, F. (2017). The role of omics in the application of adverse outcome pathways for chemical risk assessment. Toxicological Sciences, 158(2), 252–262. https://doi.org/10.1093/toxsci/kfx097
- Darde, T. A., Chalmel, F., & Svingen, T. (2018). Exploiting advances in transcriptomics to improve on human-relevant toxicology. In Current Opinion in Toxicology (Vols. 11–12, pp. 43–50). Elsevier B.V. https://doi.org/10.1016/j.cotox.2019.02.001
- Farmahin, R., Williams, A., Kuo, B., Chepelev, N. L., Thomas, R. S., Barton-Maclaren, T. S., Curran, I. H., Nong, A., Wade, M. G., & Yauk, C. L. (2017). Recommended approaches in the application of toxicogenomics to derive points of departure for chemical risk assessment. Archives of Toxicology, 91(5), 2045–2065.
- Grimm, D. (2011). The dose can make the poison: Lessons learned from adverse in vivo toxicities caused by RNAi overexpression. In Silence (Vol. 2, Issue 1). Silence. https://doi.org/10.1186/1758-907X-2-8
- Liu, Z., Huang, R., Roberts, R., & Tong, W. (2019). Toxicogenomics: A 2020 Vision. In Trends in Pharmacological Sciences (Vol. 40, Issue 2, pp. 92–103). Elsevier Ltd. https://doi.org/10.1016/j.tips.2018.12.001
- Pawar, G., Madden, J. C., Ebbrell, D., Firman, J. W., & Cronin, M. T. D. (2019). In silico toxicology data resources to support read-across and (Q)SAR. In Frontiers in Pharmacology (Vol. 10, Issue JUN, p. 561). Frontiers Media S.A. https://doi.org/10.3389/fphar.2019.00561